LAS LEGUMINOSAS EN LA AGRICULTURA DEL FUTURO / GRAIN LEGUMES IN FUTURE AGRICULTURE
HISTORY OF THE COMMON BEAN CROP: ITS EVOLUTION BEYOND ITS AREAS OF ORIGIN AND DOMESTICATION
Antonio M. De Ron
Consejo Superior de Investigaciones Científicas
amderon@mbg.csic.es
Ana M. González
Consejo Superior de Investigaciones Científicas
amgonzalez@mbg.csic.es
A. Paula Rodiño
Consejo Superior de Investigaciones Científicas
aprodino@mbg.csic.es
Marta Santalla
Consejo Superior de Investigaciones Científicas
msantalla@mbg.csic.es
Luis Godoy
Consejo Superior de Investigaciones Científicas
lgodoy@mbg.csic.es
Roberto Papa
Università Politecnica delle Marche, Ancona
rpapa@univpm.it
ABSTRACT
The common bean (Phaseolus vulgaris L.) is the most important grain legume for direct human consumption on a global scale. Current bean germplasm collections show a wide variation of phenotypes, although genetic erosion is gradually affecting this species as in many countries local traditional varieties are being replaced by elite cultivars. This crop has spread to every continent over the past few centuries, which has resulted in a complex genetic structure of bean germplasm outside its areas of origin and domestication (South and Central America). Some evidence indicates that this germplasm is more complex than previously thought and contains additional, as yet unexplored, diversity. This is especially the case in southern Europe, particularly in the Iberian Peninsula, where it was introduced in the early sixteenth century and has been documented as a secondary focus of domestication of the species. The integration of omic data into bean germplasm documentation databases and its combination with genotypic, phenotypic and agro-ecological data is opening a new era for the enhancement and efficient use of common bean genetic resources as the main grain legume in Europe and worldwide.
HISTORIA DEL CULTIVO DE LA JUDÍA: SU EVOLUCIÓN MÁS ALLÁ DE LAS ÁREAS DE ORIGEN Y DOMESTICACIÓN
RESUMEN
La judía común (Phaseolus vulgaris L.) es la leguminosa de grano más relevante para el consumo humano directo en escala global. Las colecciones de germoplasma de judía actuales muestran una amplia variación de fenotipos, aunque en muchos países las variedades locales están siendo reemplazados por cultivares de élite, concentrando la producción agraria en un número cada vez más reducido de cultivares con la consecuente erosión genética o pérdida de biodiversidad. Este cultivo se ha extendido por todos los continentes durante los últimos siglos, lo que ha dado lugar a una compleja estructura genética fuera de sus áreas de origen y domesticación (Mesoamérica y Sudamérica). Diversas evidencias indican que el germoplasma europeo contiene una diversidad adicional mayor de la esperada especialmente en el Sur de Europa, y particularmente en la Península Ibérica, dónde fue introducida a comienzos del siglo XVI, y que ha sido documentada como un centro de domesticación secundaria de la especie. La integración de datos ómicos en las bases de datos de documentación del germoplasma de judía y su combinación con datos genéticos, fenotípicos y agro-ecológicos está abriendo una nueva era para la valorización y el uso eficiente de los recursos genéticos de la judía común como la principal leguminosa de grano para consumo humano en Europa y globalmente.
Received: 08-03-2016; Accepted: 06-05-2016.
Cómo citar este artículo/Citation: De Ron, A.M.; González, A.M.; Rodiño, A.P.; Santalla, M.; Godoy, L. and Papa, R. (2016). History of the common bean crop: its evolution beyond its areas of origin and domestication. Arbor, 192 (779): a317. doi: http://dx.doi.org/10.3989/arbor.2016.779n3007
PALABRAS CLAVE: biodiversidad; domestication; evolución; genética; mejora genética; Phaseolus vulgaris.
KEYWORDS: biodiversity; breeding; domestication; evolution; genetics; Phaseolus vulgaris.
Copyright: © 2016 CSIC. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) Spain 3.0.
CONTENIDOS
THE COMMON BEAN CROP Top
The common bean (Phaseolus vulgaris L.) is the third most important food legume crop worldwide, surpassed only by soybean (Glycine max (L.) Merr.) and peanut (Arachis hypogea L.), and it is the first one for direct human consumption. Beans are produced and consumed mainly as a dry food legume, due to the high protein content of the grain, but the use of the fresh pod (snap bean) is common in many countries. Common bean is highly preferred in many parts of Africa and Latin America (where it can be the most important source of dietary protein), as well as in traditional diets of the Middle East and the Mediterranean region (Broughton et al., 2003Broughton, W. J., Hernández, G., Blair, M., Beebe, S., Gepts, P. y Vanderleyden, J. (2003). Beans (Phaseolus spp.) - model food legumes. Plant Soil, 252, pp. 55-128, http://dx.doi.org/10.1023/A:1024146710611.; Casquero, Lema, Santalla and De Ron, 2006Casquero, P. A., Lema, M., Santalla, M. y De Ron, A. M. (2006). Performance of common bean landraces from Spain in the Atlantic and Mediterranean environments. Genetic Resources Crop Evolution, 53, pp. 1021-1032, http://dx.doi.org/10.1007/s1072200477941.). This legume is part of the healthy diet of the Mediterranean basin and gaining importance in the USA where consumption has been increasing due to greater interest in “ethnic” and healthy foods (Blair and Izquierdo, 2012Blair, M. W. y Izquierdo, P. (2012). Use of the advanced backcross-QTL method to transfer seed mineral accumulation nutrition traits from wild to Andean cultivated common beans. Theoretical Applied Genetics, 125, pp. 1015-1031, http://dx.doi.org/10.1007/s001220121891x.). Recently the role of bean in human diet is being focused not only in its protein content but in the functional properties also and some authors have reported that its consumption could contribute to reduce risk of obesity, diabetes, cardiovascular diseases and colon, prostate and breast cancer (Hangen and Bennink, 2003Hangen, L. y Bennik, M. R. (2003). Consumption of black beans and navy beans (Phaseolus vulgaris) reduced azoxymethane-induced colon cancer in rats. Nutrition Cancer, 44, pp. 60-65.; Thompson, Brick, McGinley and Thompson, 2009Thompson, M. D., Brick, M. A, McGinley, J. N. y Thompson, H. J. (2009). Chemical composition and mammary cancer inhibitory activity of dry bean. Crop Science, 49, pp. 179-186, http://dx.doi.org/10.2135/cropsci2008.04.0218.). These health benefits could be due to the fiber content in the grain but also to antioxidant compounds as the phenolic ones.
DIVERSITY IN THE BEAN GERMPLASM: MARKET CLASSES Top
Common bean is extremely diverse crop in terms of cultivation methods, uses, range of environments to which has been adapted and morphological variability. It is found from sea-level up to 3,000 m. It is grown in monoculture, in association or rotation. It is used as green pods, green beans, and dry beans, even in places such as Africa young leaves are consumed as a source of vitamin A and in parts of South America roasted seeds are consumed, receiving the name “nuña” or popbean (González et al., 2014González, A. M., Yuste-Lisbona, F. J., Lores, M., De Ron, A. M., Lozano, R. y Santalla, M. (2014). Dissecting the genetic basis of popping ability in nuña bean, an ancient cultivar of common bean. Euphytica, 196, pp. 349-363, http://dx.doi.org/10.1007/s1068101310393.).
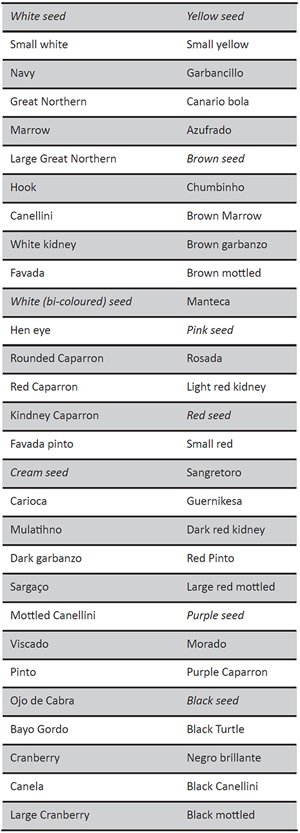
Bean markets and consumers in different countries and regions show particular preferences for grain size, shape, color and cooking time. Among some 600 bean varieties grown in the world, 62 dry bean market classes are recognized (FAO, 2002Food and Agriculture Organization of the United Nations (FAO) (2002). Production Yearbook 2002 (vol. 56). Roma: FAO.) according to consumer preferences, production and market price. Some authors have described the major worldwide market classes (Voysest, 2000Voysest, O. (2000). Mejoramiento genético del frijol (Phaseolus vulgaris L.). Legado de variedades de América Latina 1930-1999. Cali: CIAT.; Santalla, De Ron and Voyset, 2001Santalla, M., De Ron, A. M. y Voysest, O. (2001). European bean market classes. En Amurrio, M., Santalla, M. y De Ron, A. M. (eds.), Catalogue of bean genetic resources, pp. 77-94. Pontevedra: Consejo Superior de Investigaciones Científicas – Misión Biológica de Galicia, http://digital.csic.es/handle/10261/102098.) according to consumer preferences, production and market price. The main international market classes are displayed in the table 1, including some local types. Market classes usually include improved germplasm and thus tend to show a low level of variability. Breeding for commercial varieties in common bean usually occurs within each market class in order to retain their preferred seed size, shape, color, and pattern. However, the range of commercially available bean cultivars and varieties in different market classes is constantly changing. New cultivars are being released for their increased yield potential, disease resistance and improved grain.
The polymorphism of common bean is so great that, in each region, and even in each locality, different varieties with similar characteristics correspond to different names. There are several ethnic varieties or “heirloom” varieties, which are characteristic of an area or region, and they can be designated with different names. This germplasm has derived from ancient types by conscious or unconscious selection by farmers and are currently adapted to the agroecological conditions under which they have been grown for centuries.
|
Table 1. Main international common bean market classes (adapted from Santalla et al., 2001Santalla, M., De Ron, A. M. y Voysest, O. (2001). European bean market classes. En Amurrio, M., Santalla, M. y De Ron, A. M. (eds.), Catalogue of bean genetic resources, pp. 77-94. Pontevedra: Consejo Superior de Investigaciones Científicas – Misión Biológica de Galicia, http://digital.csic.es/handle/10261/102098.)
[Descargar tamaño completo]
| |
ORIGIN AND DOMESTICATION OF THE SPECIES Top
Understanding the effects of domestication on genetic diversity of common bean is of great importance, not only for crop evolution but also for possible applications, such as the implementation of appropriate biodiversity conservation strategies, and the use of genetic variability in breeding programs. One of the most important and generalised features of plant domestication is the reduction in genetic diversity, not only during the initial domestication process but also during dispersion and adaptive radiation from the centres of domestication to other areas. The reduction of genetic diversity is usually more drastic in autogamous species as common bean, which have restricted genetic recombination and present a higher population structure as compared with allogamous species (Jarvis and Hodgkin, 1999Jarvis, D. I. y Hodgkin, T. (1999). Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agro-ecosystem. Molecular Ecology, 8, pp. S159-S173, http://dx.doi.org/10.1046/j.1365294X.1999.00799.x.). This reduction is caused by both stochastic events (for example, a bottleneck and genetic drift due to a reduction in the population size) and selection (for example, for adaptation to a novel cultivated environment) (Vigouroux et al., 2002Vigouroux, Y., MacMullen, M., Hittinger, C.T., Houchins, K., Schulz, L., Kresovich, S., Matsuoka, Y. y Doebley, J. (2002). Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proceedings National Academy Sciences USA, 99, pp. 9650-9655.).
European arrival to the Americas, from the 15th century, marked the entry into Europe of a number of plant species such as common bean (Phaseolus vulgaris L.), peanuts (Arachis hypogaea L.), cocoa (Theobroma cacao L.), corn (Zea mays L.), potato (Solanum tuberosum L.) and tomato (Solanum lycopersicum L.), previously unknown in the Old World. The introduction of exotic species in a new agricultural area under different environmental conditions raises relevant questions about adaptation, taking into account the requirements of tolerance to several stresses, as well as competitiveness with other indigenous crops in production and economic value.
The taxonomic enclave of the common bean is (Sitte et al., 2004Sitte, P., Weiler, E. W., Kadereit, J. W., Bresisnky, A. y Kömer, C. (2004). Strasburger. Tratado de Botánica (35ª ed.). Barcelona: Omega.):
Class: Dicotyledoneae
Subclass: Rosidae
Superorder: Fabanae
Order: Fabales
Family: Fabaceae
Subfamily: Papilionoidae
Tribe: Phaseoleae
Subtribe: Phaseolinae
Genus: Phaseolus
Until now, over 400 species have been described in the genus Phaseolus (Freytag and Debouck, 2002Freytag, G. F. y Debouck, D. G. (2002). Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae - Papilionoideae) in North America, Mexico, and Central America. Sida, Botanical Miscellany, 23, pp. 1-300.) of which five have been domesticated and are being currently cultivated: Phaseolus vulgaris L. (common bean), P. lunatus L. (lima bean), P. coccineus L. (runner or scarlet bean), P. acutifolius A. Gray (tepary bean), and P. polyanthus Greenman (year bean). These species have genetic and phenotypic differences, particularly the reproductive system, which could be self-pollinated or outcrossed based.
Within the genus Phaseolus there are different groups or natural gene pools (Gepts and Debouck, 1991Gepts, P. y Debouck, D. (1991). Origin, domestication, and evolution of common bean (PhaseoIus vulgaris L.). En van Schoonhoven, A. y Voyest, O. (eds.), Common beans: research for crop improvement, pp. 7-53. Wallingford and Cali: CIAT.). The primary gene pool of the common bean includes the wild populations and the cultivated varieties of the species, which can intersect each other and recombine without any genetic barrier. The secondary gene pool includes the runner bean and the year bean. The crossing between common bean and the species of the secondary gene pool is easily done without embryo rescue, although using runner bean as female parent requires usually in vitro embryo rescue techniques (Bannerot, 1979Bannerot, H. (1979). Cold tolerance in beans. Annual Report Bean Improvement Cooperative, 22, pp. 81-84.). The tertiary gene pool includes the tepary bean and the crosses with common bean require techniques “in vitro. Lima bean belongs to the quaternary gene pool, and no successful crosses between the two species have been reported.
The distribution of wild common bean from northern Mexico to northwestern Argentina encompasses over 8000 km (Toro, Tohme and Debouck, 1990Toro, O., Tohme, J. y Debouck, D. G. (1990). Wild bean (Phaseolus vulgaris L.): description and distribution. Cali: CIAT.; Freytag and Debouck, 2002Freytag, G. F. y Debouck, D. G. (2002). Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae - Papilionoideae) in North America, Mexico, and Central America. Sida, Botanical Miscellany, 23, pp. 1-300.). Two major ecogeographical gene pools with partial reproductive isolation between them are recognized: Mesoamerica (from northern Mexico to Colombia) and the Andes (from southern Peru to northwestern Argentina). Additional structure within each of these gene pools accounts for ecogeographic races in each gene pool (Singh, Gepts and Debouck, 1991Singh, S. P., Gepts, P. y Debouck, D. G. (1991). Races of common bean (Phaseolus vulgaris, Fabaceae). Economic Botany, 45, pp. 379-396, http://dx.doi.org/10.1007/BF02887079.; Díaz and Blair, 2006Diaz, L. M. y Blair, M. W. (2006). Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theoretical Applied Genetics, 114, pp. 143-154, http://dx.doi.org/10.1007/s0012200604179.). Until recently, the most credited origin of the species was the northern Peru-Ecuador hypothesis (Kami, Becerra Velásquez, Debouck and Gepts, 1995Kami, J., Becerra Velásquez, B., Debouck, D. G. y Gepts, P. (1995). Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proceedings National Academy Sciences USA, 92, pp. 1101-1104, http://dx.doi.org/10.1073/pnas.92.4.1101.). Based on a DNA sequence analysis of the genes for phaseolin seed protein the authors reported that the phaseolin type I gene found in this area does not have the tandem direct repeats that are, instead, characteristic of the Mesoamerican and Andean phaseolin types. Considering that duplications, which generate tandem directs repeats, are more likely to occur than deletions, which specifically eliminate a member of a tandem direct repeat, Kami et al. (1995Kami, J., Becerra Velásquez, B., Debouck, D. G. y Gepts, P. (1995). Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proceedings National Academy Sciences USA, 92, pp. 1101-1104, http://dx.doi.org/10.1073/pnas.92.4.1101.) suggested that P. vulgaris originated from the wild populations of northern Peru and Ecuador, and subsequently spread northwards (from Colombia to northern Mexico) and southwards (from southern Peru to Argentina).
One recent alternative hypothesis for the origin of the common bean defended a Mesoamerican origin (Bitochi et al., 2012Bitocchi, E., Nanni, L., Bellucci, E., Rossi, M., Giardini, A., Zeuli, P. S., Logozzo, G., Stougaards, J., McClean, P., Attene, G. y Papa, R. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proceedings National Academy Sciences USA, 109, pp. E788-E796, http://dx.doi.org/10.1073/pnas.1108973109.; Bitochi et al., 2013Bitocchi, E., Bellucci, E., Giardini, G., Rau, R., Rodríguez, M., Biagetti, E., Santilocchi, R., Spagnoletti Zeuli, P.,, Gioia, T., Logozzo, G., Attene, G., Nanni, L. y Papa, R. (2013). Molecular analysis of the parallel domestication of the common bean in Mesoamerica and the Andes. New Phytologist, 197, pp. 300-313, http://dx.doi.org/10.1111/j.14698137.2012.04377.x.). This was mainly based on the extensive diversity and population structure within the Mesoamerican gene pool, and the signature of predomestication bottlenecks in the south of the Andes detected in five gene fragments across 102 wild accessions. This novel structure of population not only evidences a Mesoamerican origin, but also excludes an Andean origin of common bean. Additionally, these authors suggested that the wild common bean from northern Peru and Ecuador represents an old relict germplasm including a part of the genetic diversity of the ancestral common bean populations, displaying a type I phaseolin that probably was extinct in Mesoamerica. The re-sequencing of the genome of the common bean by Schmutz et al. (2014Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713, http://dx.doi.org/10.1038/ng.3008.) recently confirmed this hypothesis.
Domestication took place after the formation of these gene pools, and thus their structure is evident in both the wild and the domesticated forms (Papa and Gepts, 2003Papa, R. y Gepts, P. (2003). Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theoretical Applied Genetics, 106, pp. 239-250.; Papa, Acosta, Delgado-Salinas and Gepts, 2005Papa, R., Acosta, J., Delgado-Salinas, A. y Gepts, P. A. (2005). A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theoretical Applied Genetics, 111, pp. 1147-1158, http://dx.doi.org/10.1007/s0012200500459.; Papa et al., 2007Papa, R., Belluci, E., Rossi, M., Leonardi, S., Rau, D., Gepts, P., Nanni, L. y Attene, G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Annals Botany, 100, pp. 1039-1051, http://dx.doi.org/10.1093/aob/mcm151.; Rossi et al., 2009Rossi, M., Bitocchi, E., Belluci, E., Nanni, L., Rau, D., Attene, G. y Papa, R. (2009). Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evolutionary Applications, 2, pp. 504-522, http://dx.doi.org/10.1111/j.17524571.2009.00082.x.). This clear subdivision of the common bean germplasm is well documented, and it has been defined through several studies (Papa et al., 2007Papa, R., Belluci, E., Rossi, M., Leonardi, S., Rau, D., Gepts, P., Nanni, L. y Attene, G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Annals Botany, 100, pp. 1039-1051, http://dx.doi.org/10.1093/aob/mcm151.; Angioi et al., 2009Angioi, S. A., Rau, D., Rodriguez, M., Logozzo, G., Desiderio, F., Papa, R. y Attene, G. (2009). Nuclear and chloroplast microsatellite diversity in Phaseolus vulgaris L. from Sardinia (Italy). Molecular Breeding, 23, pp. 413-429, http://dx.doi.org/10.1007/s1103200892458.; Bitocchi et al., 2012Bitocchi, E., Nanni, L., Bellucci, E., Rossi, M., Giardini, A., Zeuli, P. S., Logozzo, G., Stougaards, J., McClean, P., Attene, G. y Papa, R. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proceedings National Academy Sciences USA, 109, pp. E788-E796, http://dx.doi.org/10.1073/pnas.1108973109.; Bitocchi et al., 2013Bitocchi, E., Bellucci, E., Giardini, G., Rau, R., Rodríguez, M., Biagetti, E., Santilocchi, R., Spagnoletti Zeuli, P.,, Gioia, T., Logozzo, G., Attene, G., Nanni, L. y Papa, R. (2013). Molecular analysis of the parallel domestication of the common bean in Mesoamerica and the Andes. New Phytologist, 197, pp. 300-313, http://dx.doi.org/10.1111/j.14698137.2012.04377.x.). However, the number of domestication events within each pool is still debated. Bitochi et al. (2013Bitocchi, E., Bellucci, E., Giardini, G., Rau, R., Rodríguez, M., Biagetti, E., Santilocchi, R., Spagnoletti Zeuli, P.,, Gioia, T., Logozzo, G., Attene, G., Nanni, L. y Papa, R. (2013). Molecular analysis of the parallel domestication of the common bean in Mesoamerica and the Andes. New Phytologist, 197, pp. 300-313, http://dx.doi.org/10.1111/j.14698137.2012.04377.x.) hypothesized a single domestication event within each gene pool and indicated the Oaxaca valley in Mesoamerica and southern Bolivia and northern Argentina as geographical areas of common bean domestication.
EVOLUTION OF THE CROP BEYOND ITS AREAS OF ORIGIN AND DOMESTICATION Top
The review of many herbaria and germplasm collections showed that so much variability exists in the genus Phaseolus. For centuries farmers have maintained their traditional or heirloom varieties and have exchanged their seeds with surrounding areas, mainly in local markets, and among themselves also. This results in a very different set of characteristics for size, shape, tenderness and cooking quality of the edible parts of the plant (pod and grain). Therefore, the traditional varieties are a valuable source of well adapted germplasm of common bean as displayed in the current common bean germplasm collections that show a wide phenotypic, although in developed countries the traditional varieties are being replaced by improved cultivars, therefore the genetic erosion is affecting the species. Deepening the phenotypic and genotypic characteristics of the different varieties in collections and genebanks is a first step to understand the variability that is currently available. This is essential to carry out breeding programs in kind, to obtain new improved varieties with desirable traits in terms of quality and production, as well as resistance to abiotic and biotic stresses.
No records of common bean earlier than 1543 have been found in European herbariums; however, as reported by Zeven (1997Zeven, A. C. (1997). The introduction of the common bean (Phaseolus vulgaris L.) into Western Europe and the phenotypic variation of dry beans collected in The Netherlands in 1946. Euphytica, 94, pp. 319-328, http://dx.doi.org/10.1023/A:1002940220241.), in 1669 it was widely grown in many areas of Europe. The dispersion of the common bean to Europe probably started from the Iberian Peninsula (Spain and Portugal), where the species was introduced mainly from Central America around 1506 (Ortwin-Sauer, 1966Ortwin-Sauer, C. (1966). The early Spanish men. Berkeley and Los Angeles: University of California Press.) and from the southern Andes after 1532, through sailors and traders who brought with them the nicely coloured and easily transportable seeds as a curiosity (Brücher and Brücher, 1976Brücher, O. B. y Brücher, H. (1976). The South American wild bean (Phaseolus aborigineus ‘Burk’.), as ancestor of the common bean. Economic Botany, 30, pp. 257-272, http://dx.doi.org/10.1007/BF02909734.; Debouck and Smartt, 1995Debouck, D. G. y Smartt, J. (1995). Beans. En Smartt, J. y Simmonds, N. W. (eds.), Evolution of Crop Plants, pp. 287-296. London: Longman Scientific & Technical.).
The common bean germplasm currently grown in the Iberian Peninsula suggests interesting questions about the nature of the variation observed, as well as the evolutionary forces affecting the current European germplasm of this crop. The Mesoamerican beans arrived in the Iberian Peninsula probably displayed limited genetic variation, represented by a small population size (population bottlenecks), and further establishment of new populations were based on a few individuals (founding events) based on farmers preferences, that could have increased genetic drift. However, later germplasm introductions from the southern Andes after around 1532, principally from Peru, could have broadened the genetic diversity (Brücher and Brücher, 1976Brücher, O. B. y Brücher, H. (1976). The South American wild bean (Phaseolus aborigineus ‘Burk’.), as ancestor of the common bean. Economic Botany, 30, pp. 257-272, http://dx.doi.org/10.1007/BF02909734.). The pathways of dissemination of the crop across Europe were very complex, with several introductions from America combined with direct exchanges between European and other Mediterranean countries (Papa et al., 2007Papa, R., Belluci, E., Rossi, M., Leonardi, S., Rau, D., Gepts, P., Nanni, L. y Attene, G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Annals Botany, 100, pp. 1039-1051, http://dx.doi.org/10.1093/aob/mcm151.). Over time, the dissemination across Europe surely occurred through seed exchanges among farmers being facilitated by territorial contiguity and similarity of environments. The protein marker phaseolin was used as a marker in describing the worldwide dissemination of common bean (Gepts, 1988Gepts, P. A. (1988). Middle American and Andean gene pool. En Gepts, P. A. (ed.), Genetic resources of Phaseolus beans, pp. 375-390. Dordrech: Kluwer, http://dx.doi.org/10.1007/9789400927865_15.). A higher frequency of Andean types (T, C, H and A) was recorded with respect to Mesoamerican ones (S, B, M) (Lioi, 1989Lioi, L. (1989). Varition in the storage protein phaseolin in common bean (Phaseolus vulgaris L.) from the Mediterranean area. Euphytica, 44, pp. 151-155, http://dx.doi.org/10.1007/BF00022610.; Santalla, Rodiño and De Ron, 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.).
Occasional outcrossing, adaptation to particular environments (in terms of temperature, moisture, photoperiod, soil fertility, diseases and insects), different cropping systems and strong selection for consumer preferences addressed to particular seed types, might have played a significant role in the evolution of new genetic variation in common bean in Europe. As a consequence, each country selected its own set of landraces able to respond to the needs and preferences of local populations. The common bean populations were involved in new evolutionary pathways that were not possible in the American center of origin, due to the spatial isolation between these two gene pools. Thus, new germplasm (e.g. favada, hook and large-great-northern class cultivars) could have arose from recombination events between Mesoamerican and Andean gene pools, better adapted to the conditions of the new agrosystems (Santalla et al., 2001Santalla, M., De Ron, A. M. y Voysest, O. (2001). European bean market classes. En Amurrio, M., Santalla, M. y De Ron, A. M. (eds.), Catalogue of bean genetic resources, pp. 77-94. Pontevedra: Consejo Superior de Investigaciones Científicas – Misión Biológica de Galicia, http://digital.csic.es/handle/10261/102098.). Evidence of this phenomenon has been detected using phaseolins, allozymes and morphological data (Santalla et al., 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.; Rodiño, Santalla, González, De Ron and Singh, 2006Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546, http://dx.doi.org/10.2135/cropsci2006.02.0104.), and ISSRs and SSRs from both the chloroplast and nuclear genomes (Sicard, Nanni, Porfiri, Bulfon and Papa, 2005Sicard, D., Nanni, L., Porfiri, O., Bulfon, D. y Papa, R. (2005). Genetic diversity of Phaseolus vulgaris L. and Phaseolus coccineus L. landraces in central Italy. Plant Breeding, 124, pp. 464-472, http://dx.doi.org/10.1111/j.14390523.2005.01137.x.; Angioi et al., 2009Angioi, S. A., Rau, D., Rodriguez, M., Logozzo, G., Desiderio, F., Papa, R. y Attene, G. (2009). Nuclear and chloroplast microsatellite diversity in Phaseolus vulgaris L. from Sardinia (Italy). Molecular Breeding, 23, pp. 413-429, http://dx.doi.org/10.1007/s1103200892458.). Gene flow between both gene pools appears to be relatively common in the Andean (Debouck, Araya Villalobos, Ocampo Sánchez and González, 1989Debouck, D. G., Araya Villalobos, R., Ocampo Sánchez, R. A. y González, U. W. G. (1989). Collecting Phaseolus in Costa Rica. Plant Genetic Resources Newsletter, 78, pp. 44-46.; Paredes and Gepts, 1995Paredes, O. M. y Gepts, P. (1995). Extensive introgression of Middle American germplasm into Chilean common bean landraces. Genetic Resources Crop Evolution, 42, pp. 9-41, http://dx.doi.org/10.1007/BF02310681.; Beebe, Toro, González, Chacón and Debouck, 1997Beebe, S., Toro, O., González, A. V., Chacón, M. I. y Debouck, D. (1997). Wildweed-crop complex of common bean (Phaseolus vulgaris L., Fabaceae) in the Andes of Peru and Colombia, and their implications for conservation and breeding. Genetic Resources Crop Evolution, 44, pp. 73-91, http://dx.doi.org/10.1023/A:1008621632680.; Chacón, Pickersgill and Debouck, 2005Chacón, M. I., Pickersgill, S. y Debouck, D. (2005). Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of Mesoamerican and Andean cultivated races. Theoretical Applied Genetics, 110, pp. 432-444, http://dx.doi.org/10.1007/s0012200418422.) and European zones (Santalla et al., 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.; Sicard et al., 2005Sicard, D., Nanni, L., Porfiri, O., Bulfon, D. y Papa, R. (2005). Genetic diversity of Phaseolus vulgaris L. and Phaseolus coccineus L. landraces in central Italy. Plant Breeding, 124, pp. 464-472, http://dx.doi.org/10.1111/j.14390523.2005.01137.x.; Piergiovanni, Taranto, Losavio and Pignone, 2006Piergiovanni, A., Taranto, G., Losavio, F. P. y Pignone, D. (2006). Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genetic Resources Crop Evolution, 53, pp. 313-322, http://dx.doi.org/10.1007/s1072200461447.; Rodiño et al., 2006Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546, http://dx.doi.org/10.2135/cropsci2006.02.0104.; Sánchez, Sifres, Casañas and Nuez, 2008Sánchez, E., Sifres, A., Casañas, F. y Nuez, F. (2008). The endangered future of organoleptically prestigious European landraces: Ganxet bean (Phaseolus vulgaris L.) as an example of a crop originating in the Americas. Genetic Resources Crop Evolution, 55, pp. 45-52, http://dx.doi.org/10.1007/s107220079213x.).
These new forms could have subsequently been disseminated to other parts of Europe, contributing to much-wider variation observed in the current European germplasm (Lioi, 1989Lioi, L. (1989). Varition in the storage protein phaseolin in common bean (Phaseolus vulgaris L.) from the Mediterranean area. Euphytica, 44, pp. 151-155, http://dx.doi.org/10.1007/BF00022610.; Gil and De Ron, 1992Gil, J. y De Ron, A. M. (1992). Variation in Phaseolus vulgaris in the northwest of the Iberian Peninsula. Plant Breeding, 109, pp. 313-319, http://dx.doi.org/10.1111/j.14390523.1992.tb00190.x.; Escribano, De Ron and Amurrio, 1994Escribano, M. R., De Ron, A. M. y Amurrio, J. M. (1994). Diversity in agronomical traits in common bean populations from Northwestern Spain. Euphytica, 76, pp. 1-6, http://dx.doi.org/10.1007/BF00024014.; Limongelli, Laghetti, Perrino, and Piergiovanni, 1996Limongelli, G., Laghetti, G., Perrino, P. y Piergiovanni, A. R. (1996). Variation of seed storage protein in landraces of common bean (Phaseolus vulgaris L.) from Basilicata, Southern Italy. Plant Breeding, 119, pp. 513-516.; Zeven, 1997Zeven, A. C. (1997). The introduction of the common bean (Phaseolus vulgaris L.) into Western Europe and the phenotypic variation of dry beans collected in The Netherlands in 1946. Euphytica, 94, pp. 319-328, http://dx.doi.org/10.1023/A:1002940220241.; González, Monteagudo, Casquero, De Ron and Santalla, 2006González, A. M, Monteagudo, A. B, Casquero, P. A, De Ron, A. M y Santalla, M. (2006). Genetic variation and environmental effects on agronomical and commercial quality traits in the main European market classes of dry bean. Field Crops Research, 95, pp. 336-347, http://dx.doi.org/10.1016/j.fcr.2005.04.004.). Therefore, the Iberian Peninsula, mainly the north and northwest regions, could be considered as a secondary center of genetic diversity for the common bean crop, especially regarding the large white-seeded cultivars (Santalla et al., 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.; Rodiño et al., 2006Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546, http://dx.doi.org/10.2135/cropsci2006.02.0104.).
The secondary diversification of the common bean and the existence of new recombinant types between the Andean and Mesoamerican genetic pools open the door for new opportunities for the genetic improvement of the species. Breeders can cross between Mesoamerican and Andean gene pools, as well among races, although it is well known that there are constraints to the crosses between Mesoamerican and Andean germplasm due to genetic barriers [blocked cotyledon lethal (BCL), crinkle leaf dwarf (CLD) and dwarf lethal (DL)] (Singh and Gutierrez, 1984Singh, S. P. y Gutiérrez, J. A. (1984). Geographical distribution of the Dl1, and Dl2 genes causing hybrid dwarfism in P vulgaris L., their association with seed size, and their significance to breeding. Euphytica, 33, pp. 337-345, http://dx.doi.org/10.1007/BF00021130.; Hannah et al., (2007Hannah, M. A., Krämer, K. M., Geffroy, V., Kopka, J., Blair, M. W., Erban, A., Eduardo Vallejos, C., Heyer, A. G., Sanders, F. T., Millner, P. A. y Pilbeam, D.J. (2007). Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytologist, 176, pp. 537-549, http://dx.doi.org/10.1111/j.14698137.2007.02215.x.). González, Rodiño, Santalla and De Ron (2009González, A. M., Rodiño, A. P., Santalla, M. y De Ron, A. M. (2009). Genetics of intra-gene pool and inter-gene pool hybridization for seed traits in common bean (Phaseolus vulgaris L.) germplasm from Europe. Field Crops Research, 112, pp. 66-76, http://dx.doi.org/10.1016/j.fcr.2009.02.003.) reported successful interracial and interpool crosses for the development of new common bean varieties in Europe. Since the Mesoamerican germplasm usually display resistance to pathogens and some Andean varieties have high seed quality, the use of the European recombinant germplasm as “bridge parents” in interpool crosses to overcome the interpool genetic barriers provides an interesting opportunity for introgression of relevant genes in the common bean varieties currently grown in Europe. Breeding can also involve gene introgression from additional genes pools, such as the secondary and tertiary gene pools, covering a range of environments from cool moist highlands to hot semi-arid regions, and from drought periods to more wet conditions.
In the case of rhizobia symbiotic system, it is possible that migration of the species had not been parallel, so additional efforts are under way to achieve efficient symbiotic genotypes of common bean and rhizobia (Rodiño et al., 2011Rodiño, A. P., De la Fuente, M., De Ron, A. M., Lema, M. J., Drevon, J. J. y Santalla, M. (2011). Variation for nodulation and plant yield of common bean genotypes and environmental effects on the genotype expression. Plant Soil, 346, pp. 349-361, http://dx.doi.org/10.1007/s111040110823x.). As a result of plant-rhizobia co-evolution, a spectrum of compatible rhizobia has developed each specific for one or more given legume species.
Common bean originated and was domesticated in tropical highlands. This means that abiotic and biotic conditions had an influence on the development of European varieties (Rodiño et al., 2006Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546, http://dx.doi.org/10.2135/cropsci2006.02.0104.; Rodiño, Riveiro, Santalla and De Ron, 2007Rodiño, A. P., Riveiro, M., Santalla, M., Santalla, M. y De Ron, A. M. (2007). Sources of variation of common bean for drought tolerance. Annual Report Bean Improvement Cooperative, 50, pp. 163-164.). In some cases, bean breeders have had to incorporate tolerances to abiotic stresses from sources outside the primary gene pool of common bean. For example, tepary bean could also provide tolerance to heat or drought, and runner bean, tolerance to low soil fertility (Miklas, Kelly, Beebe and Blair, 2006Miklas, P. N., Kelly, J. D., Beebe, S. E. y Blair, M. W. (2006). Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica, 147, pp. 106-131, http://dx.doi.org/10.1007/s1068100646005.). The disease resistance in common bean is crucial to adapt this species to new zones. The most important diseases that constrain common bean production worldwide, particularly in Europe, are anthracnose (caused by Colletotrichum lindemuthianum), rust (caused by Uromyces appendiculatus), common bacterial blight (caused by Xanthomonas axonopodis pv. phaseoli), halo blight (caused by Pseudomonas syringae pv. phaseolicola), bean common mosaic virus and bean common mosaic necrosis virus (Monteagudo et al., 2006Monteagudo, A. B., Rodiño, A. P., Lema, M., De la Fuente, M., Santalla, M., De Ron, A. M. y Singh, S. P. (2006). Resistance to fungal, bacterial and viral diseases in a common bean core collection from the Iberian Peninsula. HortScience, 41, pp. 319-322.). Pathogens causing anthracnose (Pastor-Corrales, Otoya, Molina and Singh, 1995Pastor-Corrales, M. A., Otoya, M. M., Molina, A. y Singh, S. P. (1995). Resistance to Colletotrichum lindemuthianum isolates from Middle America and Andean South America in different common bean races. Plant Disease, 79, pp. 63-67, http://dx.doi.org/10.1094/PD790063.) and rust (Sandlin, Steadman, Araya and Coyne, 1999Sandlin, C. M., Steadman, J. R, Araya, C. M. y Coyne, D. P. (1999). Isolates of Uromyces appendiculatus with specific virulence to landraces of Phaseolus vulgaris of Andean origin. Plant Disease, 83, pp. 108-113, http://dx.doi.org/10.1094/PDIS.1999.83.2.108.) have co-evolved with common bean, thereby forming clearly distinct Andean and Middle American populations. Geographic divergence between both gene pools has led to co-adaptation of the two host gene pools and their respective pathogens, such that Andean isolates are more virulent on average on Andean hosts, and vice-versa. As a consequence, breeders have sought to identify resistance for Andean host lines in the Mesoamerican host gene pool and vice versa.
In the Mediterranean basin can be clearly differentiated populations probably descendants of populations of the Iberian Peninsula where there was gene flow between Mesoamerican and Andean (Santalla et al., 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.). Gene flow between both gene pools appears to be relatively common in the Andean (Debouck et al., 1989Debouck, D. G., Araya Villalobos, R., Ocampo Sánchez, R. A. y González, U. W. G. (1989). Collecting Phaseolus in Costa Rica. Plant Genetic Resources Newsletter, 78, pp. 44-46.; Paredes and Gepts, 1995Paredes, O. M. y Gepts, P. (1995). Extensive introgression of Middle American germplasm into Chilean common bean landraces. Genetic Resources Crop Evolution, 42, pp. 9-41, http://dx.doi.org/10.1007/BF02310681.; Beebe et al., 1997Beebe, S., Toro, O., González, A. V., Chacón, M. I. y Debouck, D. (1997). Wildweed-crop complex of common bean (Phaseolus vulgaris L., Fabaceae) in the Andes of Peru and Colombia, and their implications for conservation and breeding. Genetic Resources Crop Evolution, 44, pp. 73-91, http://dx.doi.org/10.1023/A:1008621632680.; Chacón et al., 2005Chacón, M. I., Pickersgill, S. y Debouck, D. (2005). Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of Mesoamerican and Andean cultivated races. Theoretical Applied Genetics, 110, pp. 432-444, http://dx.doi.org/10.1007/s0012200418422.) and European zones (Santalla et al., 2002Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944, http://dx.doi.org/10.1007/s0012200108446.; Sicard et al., 2005Sicard, D., Nanni, L., Porfiri, O., Bulfon, D. y Papa, R. (2005). Genetic diversity of Phaseolus vulgaris L. and Phaseolus coccineus L. landraces in central Italy. Plant Breeding, 124, pp. 464-472, http://dx.doi.org/10.1111/j.14390523.2005.01137.x.; Piergiovanni et al., 2006Piergiovanni, A., Taranto, G., Losavio, F. P. y Pignone, D. (2006). Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genetic Resources Crop Evolution, 53, pp. 313-322, http://dx.doi.org/10.1007/s1072200461447.; Rodiño et al., 2006Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546, http://dx.doi.org/10.2135/cropsci2006.02.0104.; Sánchez et al., 2008Sánchez, E., Sifres, A., Casañas, F. y Nuez, F. (2008). The endangered future of organoleptically prestigious European landraces: Ganxet bean (Phaseolus vulgaris L.) as an example of a crop originating in the Americas. Genetic Resources Crop Evolution, 55, pp. 45-52, http://dx.doi.org/10.1007/s107220079213x.). Evidence of hybridisation due to the presence of morphological intermediate plants is relatively weak, since it may result from either a phenotypic plasticity or a convergent evolution rather than from a gene flow. The presence of crop-specific alleles in morphologically intermediate landraces can help to provide strong evidence for a history of hybridisation. Molecular analyses in conjunction with phenotypic studies of germplasm are recommended because they provide complementary information and increase the resolving power of the genetic diversity (Singh, Gutiérrez, Molina, Urrea, and Gepts, 1991Singh, S. P., Gutiérrez, J. A., Molina, A., Urrea, C. y Gepts, P. (1991). Genetic diversity in cultivated common bean: II. Marker-based analysis of morphological and agronomic traits. Crop Science, 31, pp. 23-29, http://dx.doi.org/10.2135/cropsci1991.0011183X003100010005x.). In the Mediterranean basin the Andean populations appear to have experienced major phenomena of evolution and adaptation, as clear differences between them appear. A particular case is the white seed beans types from Turkey that seem to be phylogenetically distant from the rest of the European beans varieties, probably due to their introduction into this country through East Asia via the Silk Route (De la Fuente, De Ron, Rodiño and Santalla, 2010De la Fuente, M., De Ron, A. M., Rodiño, A. P. y Santalla, M. (2010). Evolution of the European bean from their American ancestors. Annual Report Bean Improvement Cooperative, 53, pp. 34-35.).
After the initial domestication process, the common bean crop spread across Mesoamerica and South America and, after the European exploration of the Americas, to Europe and Africa (Gepts and Bliss, 1988Gepts, P. y Bliss, F. A. (1988). Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II. Europe and Africa. Economic Botany, 42, pp. 86-104, http://dx.doi.org/10.1007/BF02859038.; Gepts, Osborn, Rashka and Bliss, 1986Gepts, P., Osborn, T., Rashka, K. y Bliss, F. (1986). Phaseolin–protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris L.): evidence for multiple centers of domestication. Economic Botany, 40, pp. 451-468, http://dx.doi.org/10.1007/BF02859659 / http://dx.doi.org/10.1007/BF02859660.; Gepts, 1988Gepts, P. A. (1988). Middle American and Andean gene pool. En Gepts, P. A. (ed.), Genetic resources of Phaseolus beans, pp. 375-390. Dordrech: Kluwer, http://dx.doi.org/10.1007/9789400927865_15.) where it was cultivated under diverse agrosystems, environmental conditions and farmer preferences.
In Asia, China is a large producer of dry beans, and is the most important producer of snap beans in the world. An analysis of 229 landraces revealed higher prevalence of the Mesoamerican type (Zhang, Blair and Wang, 2008Zhang, X., Blair, M. W. y Wang, S. (2008). Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeats markers. Theoretical Applied Genetics, 117, pp. 629-664, http://dx.doi.org/10.1007/s0012200808072.). At present, it is believed that there were only a limited number of introductions of the common bean (“caidou”) into China, which were biased towards the Mesoamerican type. In Africa, the Mesoamerican and Andean gene pools are approximately equal in frequency (Asfaw, Blair and Almekinders, 2009Asfaw, A., Blair, M. W. y Almekinders, C. (2009). Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theoretical Applied Genetics, 120, pp. 1-12, http://dx.doi.org/10.1007/s0012200911547.; Blair, González, Kimani and Butare, 2010Blair, M. W., González, L. F., Kimani, P. M. y Butare, L. (2010). Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theoretical Applied Genetics, 121, pp. 237-248, http://dx.doi.org/10.1007/s001220101305x.), even if there are striking differences between different countries due to different farmer selection preferences and the input of germplasm from national programs. In Japan, the common bean is usually called “ingen mame” and “sasage” in the Tohoku district (the northeastern parts of Japan). The crop is considered to have been introduced into Japan in the 16th century (Hoshikawa, 1981Hoshikawa, K. (1981). Ingen mame (Common bean). En Shokuyou Sakumotu (Food Crops), pp. 482-491. Tokyo: Yoken-do.), maybe from China, and now is grown widely as a garden crop, being Hokkaido (the northernmost parts of Japan) the main area of commercial production.
Portuguese and Spanish traders probably introduced common bean to Africa from the 16th century through Sofala (Mozambique), Zanzibar and Mombasa, from where it was carried to higher altitude areas of the interior by slave trading caravans and merchants (Greenway, 1945Greenway, P. (1945). The origin of some East Africa food plants. East African Agricultural Forestry Journal, 10, pp. 177-180, http://dx.doi.org/10.1080/03670074.1945.11664438.). Common bean became well established as a pulse crop in parts of Africa before the colonial era. Genetic diversity of common bean and its pathogens and linguistic evidence indicate that it became a major crop in Central African highland areas (e.g. in Rwanda and Burundi) earlier than in other parts of Africa. The crop is of significance in many African countries and most intensively grown in the Great Lakes areas of Central Africa (Wortmann, 2006Wortmann, C. S. (2006). Phaseolus vulgaris L. (common bean). Record from PROTA4U. En Brink, M. y Belay, G. (eds.), PROTA (Plant Resources of Tropical Africa / Ressources végétales de l’Afrique tropicale). Wageningen, Netherlands, Disponible en: http://www.prota4u.org/search.asp.). Many varieties of beans are grown in Africa, with wide diversity in seed types and adaptation to climatic and agronomic very different. Additionally, the consumer and market preferences have produced a high phenotypic diversity in this species. For these reasons, Africa highlands are proposed as a secondary center of diversity for the species by Asfaw et al. (2009Asfaw, A., Blair, M. W. y Almekinders, C. (2009). Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theoretical Applied Genetics, 120, pp. 1-12, http://dx.doi.org/10.1007/s0012200911547.) and Blair et al., (2010Blair, M. W., González, L. F., Kimani, P. M. y Butare, L. (2010). Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theoretical Applied Genetics, 121, pp. 237-248, http://dx.doi.org/10.1007/s001220101305x.). Blair et al. (2010Blair, M. W., González, L. F., Kimani, P. M. y Butare, L. (2010). Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theoretical Applied Genetics, 121, pp. 237-248, http://dx.doi.org/10.1007/s001220101305x.) identified Andean and Mesoamerican distinct genotypes and landraces intermediate between the gene pools and representing inter-gene pool introgression in terms of phenotypic characteristics and alleles.
BREEDING PERSPECTIVES Top
Common bean has become, over the last twenty years, in a competitive crop in national, regional and international markets. This situation presents a dynamic environment for producers and researchers of this crop and requires a rethinking of current strategies against research and production needs, the opportunities and challenges of the future.
An important long-term challenge is the discovery of the gene(s) that control important production traits. This will need to be a cooperative world-wide effort that involves breeders, geneticists, and genomic and bioinformatics experts. Breeders provide the essential skills of phenotyping, and the identification and development of genetic populations. Connecting phenotyping with the functional gene requires the skills of pathologists, physiologists, and those with a deep knowledge of plant anatomy. Those skilled with genomics and bioinformatics provide the expertise to link the phenotypic and genotypic data with candidate genes. Once a candidate gene is defined and the causative mutation is discovered, breeders will then have access to best possible marker, one that is in the gene controlling the important phenotype.
To date, considerable efforts have been made towards DNA polymorphisms discovery in common bean. Several thousand Single nucleotide polymorphisms (SNPs) and insertions-deletions (InDels) have been discovered through expressed sequence tags data mining and partial re-sequencing of several genotypes (Hyten et al., 2010Hyten, D. L, Song, Q., Fickus, E. W., Quigley, C. V., Lim, J. S., Choi, I. Y., Hwang, E. Y., Pastor-Corrales, M. y Cregan, P. B. (2010). High through-put SNP discovery and assay development in common bean. BMC Genomics, 11, pp. 475-483, http://dx.doi.org/10.1186/1471216411475.; Souza et al., 2012Souza, T. L. P. O., Barros, E. G., Bellato, C. M., Hwang, E-Y, Cregan, P. C. y Pastor-Corrales, M. A. (2012). Single nucleotide polymorphism discovery in common bean. Molecular Breeding, 30, pp. 419-428, http://dx.doi.org/10.1007/s1103201196324.; Felicetti et al., 2012Felicetti, E., Song, Q., Jia, G., Cregan, P., Bett, K. E. y Miklas, P. N. (2012). Simple sequence repeats linked with slow darkening trait in pinto bean discovered by single nucleotide polymorphism assay and whole genome sequencing. Crop Science, 52, pp. 1600-1608, http://dx.doi.org/10.2135/cropsci2011.12.0655.; Blair et al., 2013Blair, M. W., Cortes, A. J., Penmetsa, R. V., Farmer, A., Carrasquilla-García, N. y Cook, D. R. (2013). A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theoretical Applied Genetics, 126, pp. 535-548, http://dx.doi.org/10.1007/s001220121999z.; Goretti et al., 2014Goretti, D., Bitocchi, E., Bellucci, E., Rodríguez, M., Rau, D., Gioia, T., Attene, G., McClean, P., Nanni, L. y Papa, R. (2014). Development of single nucleotide polymorphisms in Phaseolus vulgaris and related Phaseolus spp. Molecular Breeding, 33, pp. 531-544, http://dx.doi.org/10.1007/s1103201399705.; Zou et al., 2014Zou, X. L., Shi, C., Austin, R. S., Merico, D., Munholland, S., Marsolais, F., Navabi, A., Crosby, W. L., Pauls, K. P., Yu, K. y Cui, Y. (2014). Genome-wide single nucleotide polymorphism and insertion-deletion discovery through next-generation sequencing of reduced representation libraries in common bean. Molecular Breeding, 33, pp. 769-778, http://dx.doi.org/10.1007/s1103201399977.). At the transcriptional level, expressed sequence tags (ESTs) sequencing has been used to discover and identify genes differentially expressed under different conditions. Whole genome transcriptome analysis is also an effective way to exploit key factors for common bean responses to biotic and abiotic stress that are involved in transcriptional and metabolic activities. The data obtained from these technologies will serve as an invaluable genomic reference to further our knowledge about the common bean at the molecular level, and can be applied to molecular breeding for plants with enhanced biotic and abiotic tolerance.
The genome of an Andean common bean genotype (G19833) was sequenced and recently released (Schmutz et al., 2014Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713, http://dx.doi.org/10.1038/ng.3008.). A combination of Sanger, 454, and Illumina HiSeq2000 reads and a genetic map based on 7015 SNP markers were used to assemble the common bean reference genome sequence (Schmutz et al., 2014Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713, http://dx.doi.org/10.1038/ng.3008.), with a total genome size of 521 Mb that represents 89% of the 587 Mb bean genome. Also, a first draft of the entire common bean genome sequence of a Mesoamerican genotype (BAT93). was also developed under the framework of the PhasIbeAm consortium within the Project “Sequencing of the common bean genome (Phaseolus vulgaris L.) for the rational exploitation of the natural resources of Iberoamerica” funded by the Ibero-American Programme for Science, Technology and Development (CYTED). The genome sequence has an immediate application by providing a reference from which new markers can be developed.
Currently new technologies built around the recently released common bean genome sequence (Schmutz et al., 2014Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713, http://dx.doi.org/10.1038/ng.3008.) are now being developed. Regarding the new breeding technologies, genetic transformation causes some public concern in many countries, but novel breeding material obtained by mutagens are more acceptable to consumers, breeders, and governments. In this context, Targeting Induced Local Lesions in Genome (TILLING) technology has been developed as a new powerful breeding methodology (De Ron et al., 2015De Ron, A. M., Papa, R., Bitocchi, E., González, A. M., Debouck, D. G., Brick, M. A., Fourie, D., Marsolais, F., Beaver, J., Geffroy, V., McClean, P., Santalla, M., Lozano, R., Yuste-Lisbona, F. J. y Casquero, P. A. (2015). Common Bean. En De Ron, A. M. (ed.), Grain Legumes, pp. 1-36. New York: Springer, http://dx.doi.org/10.1007/9781493927975_1 / http://dx.doi.org/10.1007/9781493927975 .). TILLING is a non-transgenic method that uses gene-specific primers for the identification of mutants of a gene of interest from a large mutagenesis population (McCallum, Comai, Greene and Henikoff, 2000McCallum, C. M., Comai, L., Greene, E. A. y Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiology, 123, pp. 439-442, http://dx.doi.org/10.1104/pp.123.2.439.). TILLING has gained popularity as a reverse genetic approach because it can produce series of mutants, including knockouts, and it does not rely on the transformation method for gene discovery and verification. Significant advances have been made in the development of a TILLING platform in common bean, but the protocol for this crop has yet to be optimized. Induced mutation breeding is an effective method to increase the common bean genetic variability available to the plant breeders. Additionally, renewed interest is being generated in induced mutations since the sequence of the common bean genome is already available and it will bring new opportunities for functional genomics research. Therefore, induced mutagenesis will probably become a powerful tool for the isolation and functional characterization of interesting genes, which can be used in common bean genetic improvement.
Improvement of the common bean means possessing in-depth knowledge of its genetic diversity, the genome and gene functions, to enable the analysis of pathways and networks in response to fluctuating environmental conditions. Various genomic resources for common bean are available and include physical maps, bacterial artificial chromosome libraries, anchored physical and genetic maps, expressed sequence tags, and the recently published complete genome sequence (Schmutz et al. 2014Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713, http://dx.doi.org/10.1038/ng.3008.). However, these approaches require precise phenotypic data. Complex interactions between the crop genotype, environmental factors in combination with plant population dynamics and crop management greatly affect plant phenotypes in field experiments. Hence, novel techniques should be kept cost-effective and robust under varying field conditions and should allow for the monitoring of various and complex traits.
BIBLIOGRAPHYTop
|
| ○ | Asfaw, A., Blair, M. W. y Almekinders, C. (2009). Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theoretical Applied Genetics, 120, pp. 1-12. http://dx.doi.org/10.1007/s00122-009-1154-7. |
| ○ | Angioi, S. A., Rau, D., Rodriguez, M., Logozzo, G., Desiderio, F., Papa, R. y Attene, G. (2009). Nuclear and chloroplast microsatellite diversity in Phaseolus vulgaris L. from Sardinia (Italy). Molecular Breeding, 23, pp. 413-429. http://dx.doi.org/10.1007/s11032-008-9245-8. |
| ○ | Bannerot, H. (1979). Cold tolerance in beans. Annual Report Bean Improvement Cooperative, 22, pp. 81-84. |
| ○ | Beebe, S., Toro, O., González, A. V., Chacón, M. I. y Debouck, D. (1997). Wildweed-crop complex of common bean (Phaseolus vulgaris L., Fabaceae) in the Andes of Peru and Colombia, and their implications for conservation and breeding. Genetic Resources Crop Evolution, 44, pp. 73-91. http://dx.doi.org/10.1023/A:1008621632680. |
| ○ | Bellucci, E., Bitocchi, E., Rau, D., Rodríguez, M., Biagetti, E., Giardini, A., Attene, G., Nanni, L. y Papa, R. (2014). Genomics of origin, domestication and evolution of Phaseolus vulgaris. En Tuberosa, R., Graner, A. y Frison, E. (eds.), Genomics of plant genetic resources, pp. 483-507. Dordrecht: Springer. http://dx.doi.org/10.1007/978-94-007-7572-5. |
| ○ | Bitocchi, E., Nanni, L., Bellucci, E., Rossi, M., Giardini, A., Zeuli, P. S., Logozzo, G., Stougaards, J., McClean, P., Attene, G. y Papa, R. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proceedings National Academy Sciences USA, 109, pp. E788-E796. http://dx.doi.org/10.1073/pnas.1108973109. |
| ○ | Bitocchi, E., Bellucci, E., Giardini, G., Rau, R., Rodríguez, M., Biagetti, E., Santilocchi, R., Spagnoletti Zeuli, P., Gioia, T., Logozzo, G., Attene, G., Nanni, L. y Papa, R. (2013). Molecular analysis of the parallel domestication of the common bean in Mesoamerica and the Andes. New Phytologist, 197, pp. 300-313. http://dx.doi.org/10.1111/j.1469-8137.2012.04377.x. |
| ○ | Blair, M. W., González, L. F., Kimani, P. M. y Butare, L. (2010). Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theoretical Applied Genetics, 121, pp. 237-248. http://dx.doi.org/10.1007/s00122-010-1305-x. |
| ○ | Blair, M. W. y Izquierdo, P. (2012). Use of the advanced backcross-QTL method to transfer seed mineral accumulation nutrition traits from wild to Andean cultivated common beans. Theoretical Applied Genetics, 125, pp. 1015-1031. http://dx.doi.org/10.1007/s00122-012-1891-x. |
| ○ | Blair, M. W., Cortes, A. J., Penmetsa, R. V., Farmer, A., Carrasquilla-García, N. y Cook, D. R. (2013). A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theoretical Applied Genetics, 126, pp. 535-548. http://dx.doi.org/10.1007/s00122-012-1999-z. |
| ○ | Broughton, W. J., Hernández, G., Blair, M., Beebe, S., Gepts, P. y Vanderleyden, J. (2003). Beans (Phaseolus spp.) - model food legumes. Plant Soil, 252, pp. 55-128. http://dx.doi.org/10.1023/A:1024146710611. |
| ○ | Brücher, O. B. y Brücher, H. (1976). The South American wild bean (Phaseolus aborigineus ‘Burk’.), as ancestor of the common bean. Economic Botany, 30, pp. 257-272. http://dx.doi.org/10.1007/BF02909734. |
| ○ | Casquero, P. A., Lema, M., Santalla, M. y De Ron, A. M. (2006). Performance of common bean landraces from Spain in the Atlantic and Mediterranean environments. Genetic Resources Crop Evolution, 53, pp. 1021-1032. http://dx.doi.org/10.1007/s10722-004-7794-1. |
| ○ | Chacón, M. I., Pickersgill, S. y Debouck, D. (2005). Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of Mesoamerican and Andean cultivated races. Theoretical Applied Genetics, 110, pp. 432-444. http://dx.doi.org/10.1007/s00122-004-1842-22. |
| ○ | De la Fuente, M., De Ron, A. M., Rodiño, A. P. y Santalla, M. (2010). Evolution of the European bean from their American ancestors. Annual Report Bean Improvement Cooperative, 53, pp. 34-35. |
| ○ | De Ron, A. M., Papa, R., Bitocchi, E., González, A. M., Debouck, D. G., Brick, M. A., Fourie, D., Marsolais, F., Beaver, J., Geffroy, V., McClean, P., Santalla, M., Lozano, R., Yuste-Lisbona, F. J. y Casquero, P. A. (2015). Common Bean. En De Ron, A. M. (ed.), Grain Legumes, pp. 1-36. New York: Springer. http://dx.doi.org/10.1007/978-1-4939-2797-5_1. |
| ○ | Debouck, D. G., Araya Villalobos, R., Ocampo Sánchez, R. A. y González, U. W. G. (1989). Collecting Phaseolus in Costa Rica. Plant Genetic Resources Newsletter, 78, pp. 44-46. |
| ○ | Debouck, D. G. y Smartt, J. (1995). Beans. En Smartt, J. y Simmonds, N. W. (eds.), Evolution of Crop Plants, pp. 287-296. London: Longman Scientific & Technical. |
| ○ | Diaz, L. M. y Blair, M. W. (2006). Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theoretical Applied Genetics, 114, pp. 143-154. http://dx.doi.org/10.1007/s00122-006-0417-9. |
| ○ | Escribano, M. R., De Ron, A. M. y Amurrio, J. M. (1994). Diversity in agronomical traits in common bean populations from Northwestern Spain. Euphytica, 76, pp. 1-6. http://dx.doi.org/10.1007/BF00024014. |
| ○ | Felicetti, E., Song, Q., Jia, G., Cregan, P., Bett, K. E. y Miklas, P. N. (2012). Simple sequence repeats linked with slow darkening trait in pinto bean discovered by single nucleotide polymorphism assay and whole genome sequencing. Crop Science, 52, pp. 1600-1608. http://dx.doi.org/10.2135/cropsci2011.12.0655. |
| ○ | Food and Agriculture Organization of the United Nations (FAO) (2002). Production Yearbook 2002 (vol. 56). Roma: FAO. |
| ○ | Freytag, G. F. y Debouck, D. G. (2002). Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae - Papilionoideae) in North America, Mexico, and Central America. Sida, Botanical Miscellany, 23, pp. 1-300. |
| ○ | Gepts, P., Osborn, T., Rashka, K. y Bliss, F. (1986). Phaseolin–protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris L.): evidence for multiple centers of domestication. Economic Botany, 40, pp. 451-468. http://dx.doi.org/10.1007/BF02859659 / http://dx.doi.org/10.1007/BF02859660. |
| ○ | Gepts, P. A. (1988). Middle American and Andean gene pool. En Gepts, P. A. (ed.), Genetic resources of Phaseolus beans, pp. 375-390. Dordrech: Kluwer. http://dx.doi.org/10.1007/978-94-009-2786-5_15. |
| ○ | Gepts, P. y Bliss, F. A. (1988). Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II. Europe and Africa. Economic Botany, 42, pp. 86-104. http://dx.doi.org/10.1007/BF02859038. |
| ○ | Gepts, P. y Debouck, D. (1991). Origin, domestication, and evolution of common bean (PhaseoIus vulgaris L.). En van Schoonhoven, A. y Voyest, O. (eds.), Common beans: research for crop improvement, pp. 7-53. Wallingford and Cali: CIAT. |
| ○ | Gil, J. y De Ron, A. M. (1992). Variation in Phaseolus vulgaris in the northwest of the Iberian Peninsula. Plant Breeding, 109, pp. 313-319. http://dx.doi.org/10.1111/j.1439-0523.1992.tb00190.x. |
| ○ | González, A. M, Monteagudo, A. B, Casquero, P. A, De Ron, A. M y Santalla, M. (2006). Genetic variation and environmental effects on agronomical and commercial quality traits in the main European market classes of dry bean. Field Crops Research, 95, pp. 336-347. http://dx.doi.org/10.1016/j.fcr.2005.04.004. |
| ○ | González, A. M., Rodiño, A. P., Santalla, M. y De Ron, A. M. (2009). Genetics of intra-gene pool and inter-gene pool hybridization for seed traits in common bean (Phaseolus vulgaris L.) germplasm from Europe. Field Crops Research, 112, pp. 66-76. http://dx.doi.org/10.1016/j.fcr.2009.02.003. |
| ○ | González, A. M., Yuste-Lisbona, F. J., Lores, M., De Ron, A. M., Lozano, R. y Santalla, M. (2014). Dissecting the genetic basis of popping ability in nuña bean, an ancient cultivar of common bean. Euphytica, 196, pp. 349-363. http://dx.doi.org/10.1007/s10681-013-1039-3. |
| ○ | Goretti, D., Bitocchi, E., Bellucci, E., Rodríguez, M., Rau, D., Gioia, T., Attene, G., McClean, P., Nanni, L. y Papa, R. (2014). Development of single nucleotide polymorphisms in Phaseolus vulgaris and related Phaseolus spp. Molecular Breeding, 33, pp. 531-544. http://dx.doi.org/10.1007/s11032-013-9970-5. |
| ○ | Greenway, P. (1945). The origin of some East Africa food plants. East African Agricultural Forestry Journal, 10, pp. 177-180. http://dx.doi.org/10.1080/03670074.1945.11664438. |
| ○ | Hangen, L. y Bennik, M. R. (2003). Consumption of black beans and navy beans (Phaseolus vulgaris) reduced azoxymethane-induced colon cancer in rats. Nutrition Cancer, 44, pp. 60-65. |
| ○ | Hoshikawa, K. (1981). Ingen mame (Common bean). En Shokuyou Sakumotu (Food Crops), pp. 482-491. Tokyo: Yoken-do. |
| ○ | Hannah, M. A., Krämer, K. M., Geffroy, V., Kopka, J., Blair, M. W., Erban, A., Eduardo Vallejos, C., Heyer, A. G., Sanders, F. T., Millner, P. A. y Pilbeam, D.J. (2007). Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytologist, 176, pp. 537-549. http://dx.doi.org/10.1111/j.1469-8137.2007.02215.x. |
| ○ | Hyten, D. L, Song, Q., Fickus, E. W., Quigley, C. V., Lim, J. S., Choi, I. Y., Hwang, E. Y., Pastor-Corrales, M. y Cregan, P. B. (2010). High through-put SNP discovery and assay development in common bean. BMC Genomics, 11, pp. 475-483. http://dx.doi.org/10.1186/1471-2164-11-475. |
| ○ | Jarvis, D. I. y Hodgkin, T. (1999). Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agro-ecosystem. Molecular Ecology, 8, pp. S159-S173. http://dx.doi.org/10.1046/j.1365-294X.1999.00799.x. |
| ○ | Kami, J., Becerra Velásquez, B., Debouck, D. G. y Gepts, P. (1995). Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proceedings National Academy Sciences USA, 92, pp. 1101-1104. http://dx.doi.org/10.1073/pnas.92.4.1101. |
| ○ | Limongelli, G., Laghetti, G., Perrino, P. y Piergiovanni, A. R. (1996). Variation of seed storage protein in landraces of common bean (Phaseolus vulgaris L.) from Basilicata, Southern Italy. Plant Breeding, 119, pp. 513-516. |
| ○ | Lioi, L. (1989). Varition in the storage protein phaseolin in common bean (Phaseolus vulgaris L.) from the Mediterranean area. Euphytica, 44, pp. 151-155. http://dx.doi.org/10.1007/BF00022610. |
| ○ | McCallum, C. M., Comai, L., Greene, E. A. y Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiology, 123, pp. 439-442. http://dx.doi.org/10.1104/pp.123.2.439. |
| ○ | Miklas, P. N., Kelly, J. D., Beebe, S. E. y Blair, M. W. (2006). Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica, 147, pp. 106-131. http://dx.doi.org/10.1007/s10681-006-4600-5. |
| ○ | Monteagudo, A. B., Rodiño, A. P., Lema, M., De la Fuente, M., Santalla, M., De Ron, A. M. y Singh, S. P. (2006). Resistance to fungal, bacterial and viral diseases in a common bean core collection from the Iberian Peninsula. HortScience, 41, pp. 319-322. |
| ○ | Ortwin-Sauer, C. (1966). The early Spanish men. Berkeley and Los Angeles: University of California Press. |
| ○ | Papa, R. y Gepts, P. (2003). Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theoretical Applied Genetics, 106, pp. 239-250. |
| ○ | Papa, R., Acosta, J., Delgado-Salinas, A. y Gepts, P. A. (2005). A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theoretical Applied Genetics, 111, pp. 1147-1158. http://dx.doi.org/10.1007/s00122-005-0045-9. |
| ○ | Papa, R., Belluci, E., Rossi, M., Leonardi, S., Rau, D., Gepts, P., Nanni, L. y Attene, G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Annals Botany, 100, pp. 1039-1051. http://dx.doi.org/10.1093/aob/mcm151. |
| ○ | Paredes, O. M. y Gepts, P. (1995). Extensive introgression of Middle American germplasm into Chilean common bean landraces. Genetic Resources Crop Evolution, 42, pp. 9-41. http://dx.doi.org/10.1007/BF02310681. |
| ○ | Pastor-Corrales, M. A., Otoya, M. M., Molina, A. y Singh, S. P. (1995). Resistance to Colletotrichum lindemuthianum isolates from Middle America and Andean South America in different common bean races. Plant Disease, 79, pp. 63-67. http://dx.doi.org/10.1094/PD-79-0063. |
| ○ | Piergiovanni, A., Taranto, G., Losavio, F. P. y Pignone, D. (2006). Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genetic Resources Crop Evolution, 53, pp. 313-322. http://dx.doi.org/10.1007/s10722-004-6144-7. |
| ○ | Rodiño, P., Santalla, M., González, A. M., De Ron, A. M. y Singh, S. P. (2006). Novel genetic variation in common bean from the Iberian Peninsula. Crop Science, 46, pp. 2540-2546. http://dx.doi.org/10.2135/cropsci2006.02.0104. |
| ○ | Rodiño, A. P., Riveiro, M., Santalla, M., Santalla, M. y De Ron, A. M. (2007). Sources of variation of common bean for drought tolerance. Annual Report Bean Improvement Cooperative, 50, pp. 163-164. |
| ○ | Rodiño, A. P., De la Fuente, M., De Ron, A. M., Lema, M. J., Drevon, J. J. y Santalla, M. (2011). Variation for nodulation and plant yield of common bean genotypes and environmental effects on the genotype expression. Plant Soil, 346, pp. 349-361. http://dx.doi.org/10.1007/s11104-011-0823-x. |
| ○ | Rossi, M., Bitocchi, E., Belluci, E., Nanni, L., Rau, D., Attene, G. y Papa, R. (2009). Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evolutionary Applications, 2, pp. 504-522. http://dx.doi.org/10.1111/j.1752-4571.2009.00082.x. |
| ○ | Sandlin, C. M., Steadman, J. R, Araya, C. M. y Coyne, D. P. (1999). Isolates of Uromyces appendiculatus with specific virulence to landraces of Phaseolus vulgaris of Andean origin. Plant Disease, 83, pp. 108-113. http://dx.doi.org/10.1094/PDIS.1999.83.2.108. |
| ○ | Sánchez, E., Sifres, A., Casañas, F. y Nuez, F. (2008). The endangered future of organoleptically prestigious European landraces: Ganxet bean (Phaseolus vulgaris L.) as an example of a crop originating in the Americas. Genetic Resources Crop Evolution, 55, pp. 45-52. http://dx.doi.org/10.1007/s10722-007-9213-x. |
| ○ | Santalla, M., De Ron, A. M. y Voysest, O. (2001). European bean market classes. En Amurrio, M., Santalla, M. y De Ron, A. M. (eds.), Catalogue of bean genetic resources, pp. 77-94. Pontevedra: Consejo Superior de Investigaciones Científicas – Misión Biológica de Galicia. http://digital.csic.es/handle/10261/102098. |
| ○ | Santalla, M., Rodiño, A. P. y De Ron, A. M. (2002). Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theoretical Applied Genetics, 104, pp. 934-944. http://dx.doi.org/10.1007/s00122-001-0844-6. |
| ○ | Sicard, D., Nanni, L., Porfiri, O., Bulfon, D. y Papa, R. (2005). Genetic diversity of Phaseolus vulgaris L. and Phaseolus coccineus L. landraces in central Italy. Plant Breeding, 124, pp. 464-472. http://dx.doi.org/10.1111/j.1439-0523.2005.01137.x. |
| ○ | Singh, S. P. y Gutiérrez, J. A. (1984). Geographical distribution of the Dl1, and Dl2 genes causing hybrid dwarfism in P vulgaris L., their association with seed size, and their significance to breeding. Euphytica, 33, pp. 337-345. http://dx.doi.org/10.1007/BF00021130. |
| ○ | Singh, S. P., Gepts, P. y Debouck, D. G. (1991). Races of common bean (Phaseolus vulgaris, Fabaceae). Economic Botany, 45, pp. 379-396. http://dx.doi.org/10.1007/BF02887079. |
| ○ | Singh, S. P., Gutiérrez, J. A., Molina, A., Urrea, C. y Gepts, P. (1991). Genetic diversity in cultivated common bean: II. Marker-based analysis of morphological and agronomic traits. Crop Science, 31, pp. 23-29. http://dx.doi.org/10.2135/cropsci1991.0011183X003100010005x. |
| ○ | Sitte, P., Weiler, E. W., Kadereit, J. W., Bresisnky, A. y Kömer, C. (2004). Strasburger. Tratado de Botánica (35ª ed.). Barcelona: Omega. |
| ○ | Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., Jenkins, J., Shu, S., Song, Q., Chavarro, C., Torres-Torres, M., Geffroy, V., Moghaddam, S. M., Gao, D., Abernathy, B., Barry, K., Blair, M., Brick, M. A., Chovatia, M., Gepts, P., Goodstein, D. M., Gonzales, M., Hellsten, U., Hyten, D. L., Jia, G, Kelly, J.D., Kudrna, D., Lee, R., Richard, M. M., Miklas, P. N., Osorno, J. M., Rodrigues, J., Thareau, V., Urrea, C. A., Wang, M., Yu, Y., Zhang, M., Wing, R. A., Cregan, P. B., Rokhsar, D. S. y Jackson, S. A. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics, 46, pp. 707-713. http://dx.doi.org/10.1038/ng.3008. |
| ○ | Souza, T. L. P. O., Barros, E. G., Bellato, C. M., Hwang, E-Y, Cregan, P. C. y Pastor-Corrales, M. A. (2012). Single nucleotide polymorphism discovery in common bean. Molecular Breeding, 30, pp. 419-428. http://dx.doi.org/10.1007/s11032-011-9632-4. |
| ○ | Thompson, M. D., Brick, M. A, McGinley, J. N. y Thompson, H. J. (2009). Chemical composition and mammary cancer inhibitory activity of dry bean. Crop Science, 49, pp. 179-186. http://dx.doi.org/10.2135/cropsci2008.04.0218. |
| ○ | Toro, O., Tohme, J. y Debouck, D. G. (1990). Wild bean (Phaseolus vulgaris L.): description and distribution. Cali: CIAT. |
| ○ | Vigouroux, Y., MacMullen, M., Hittinger, C.T., Houchins, K., Schulz, L., Kresovich, S., Matsuoka, Y. y Doebley, J. (2002). Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proceedings National Academy Sciences USA, 99, pp. 9650-9655. |
| ○ | Voysest, O. (2000). Mejoramiento genético del frijol (Phaseolus vulgaris L.). Legado de variedades de América Latina 1930-1999. Cali: CIAT. |
| ○ | Wortmann, C. S. (2006). Phaseolus vulgaris L. (common bean). Record from PROTA4U. En Brink, M. y Belay, G. (eds.), PROTA (Plant Resources of Tropical Africa / Ressources végétales de l’Afrique tropicale). Wageningen, Netherlands, Disponible en: http://www.prota4u.org/search.asp. |
| ○ | Zhang, X., Blair, M. W. y Wang, S. (2008). Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeats markers. Theoretical Applied Genetics, 117, pp. 629-664. http://dx.doi.org/10.1007/s00122-008-0807-2. |
| ○ | Zeven, A. C. (1997). The introduction of the common bean (Phaseolus vulgaris L.) into Western Europe and the phenotypic variation of dry beans collected in The Netherlands in 1946. Euphytica, 94, pp. 319-328. http://dx.doi.org/10.1023/A:1002940220241. |
| ○ | Zou, X. L., Shi, C., Austin, R. S., Merico, D., Munholland, S., Marsolais, F., Navabi, A., Crosby, W. L., Pauls, K. P., Yu, K. y Cui, Y. (2014). Genome-wide single nucleotide polymorphism and insertion-deletion discovery through next-generation sequencing of reduced representation libraries in common bean. Molecular Breeding, 33, pp. 769-778. http://dx.doi.org/10.1007/s11032-013-9997-7. |
|